Methods and Theory : Congealing 3D Medical Volumes
Overview
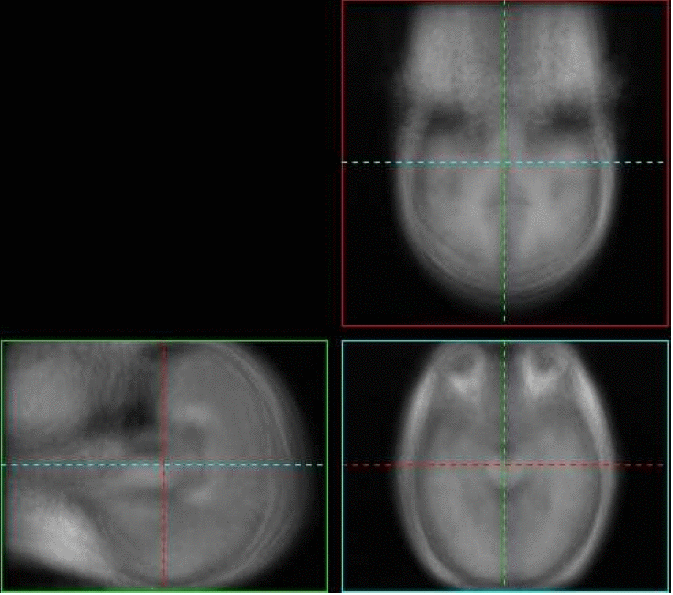
In his thesis, Learned-Miller experimented with congealing, or jointly aligning, 3D brain volumes. However, before the volumes could be aligned, the volumes had to be binarized, or thresholded to values of zero or one. While congealing was defined for 3D gray scale volumes, the method was too slow to generalize well to these large data sets. Lilla Zollei developed fast algorithms for congealing 3D MRI scans of human heads. See the paper below for more details.The images below show various sections through the average of a set of poorly aligned 3D MRI brain images. Try moving your mouse over the brain images and they will be replaced by the average of the congealed brain images. The sharpened average suggests that the congealed images are better aligned than the originals.
Faculty
Collaborators
Publications
- Lilla Zollei, Erik Learned-Miller, Eric Grimson, and William Wells.
Efficient population registration of 3D data.
In Workshop on Computer Vision for Biomedical Image Applications: Current Techniques and Future Trends, at the International Conference of Computer Vision (best paper award), 2005.
[pdf] - Erik Miller.
Learning from one example in machine vision by sharing probability densities.
Ph.D. Thesis, Massachusetts Institute of Technology, Department of Electrical Engineering and Computer Science, 2002.
[pdf] - Erik Learned-Miller.
Data driven image models through continuous joint alignment.
In IEEE Transactions on Pattern Analysis and Machine Intelligence (PAMI), 28:2, pp. 236-250, 2006.
[pdf]
